Cross-mapping biodiversity metrics across spaces
Bruno Vilela
2025-07-07
Source:vignettes/Cross-mapping-biodiversity-metrics-Phyllomedusa.Rmd
Cross-mapping-biodiversity-metrics-Phyllomedusa.RmdOverview
This vignette demonstrates cross-mapping of
biodiversity metrics between environmental,
geographic, and attribute (trait)
spaces using letsR. We:
- Build an environmental-space PAM from Phyllomedusa data;
- Compute per-cell descriptors in environmental space;
- Summarize those descriptors to species level; and
- Project (cross-map) a chosen environmental metric into geographic and attribute spaces via the package connectors.
Data and environmental PAM (example with Phyllomedusa)
First we create a geographic and environmental PAM as described in previous articles.
library(letsR)
# Occurrences and climate
data("Phyllomedusa"); data("prec"); data("temp")
prec <- terra::unwrap(prec); temp <- terra::unwrap(temp)
# Geographic PAM (keep empty cells)
PAM <- lets.presab(Phyllomedusa, remove.cells = FALSE)
# Keep terrestrial landmasses for plotting and binning consistency
wrld_simpl <- get(utils::data("wrld_simpl", package = "letsR"))
PAM <- lets.pamcrop(PAM, terra::vect(wrld_simpl))
# Environmental variables matrix (per geographic cell)
envs <- lets.addvar(PAM, c(temp, prec), onlyvar = TRUE)
colnames(envs) <- c("Temperature", "Precipitation")
# Environmental-space PAM (envPAM)
env_obj <- lets.envpam(PAM, envs, n_bins = 30, remove.cells = FALSE)
# Plot
lets.plot.envpam(env_obj, world = TRUE)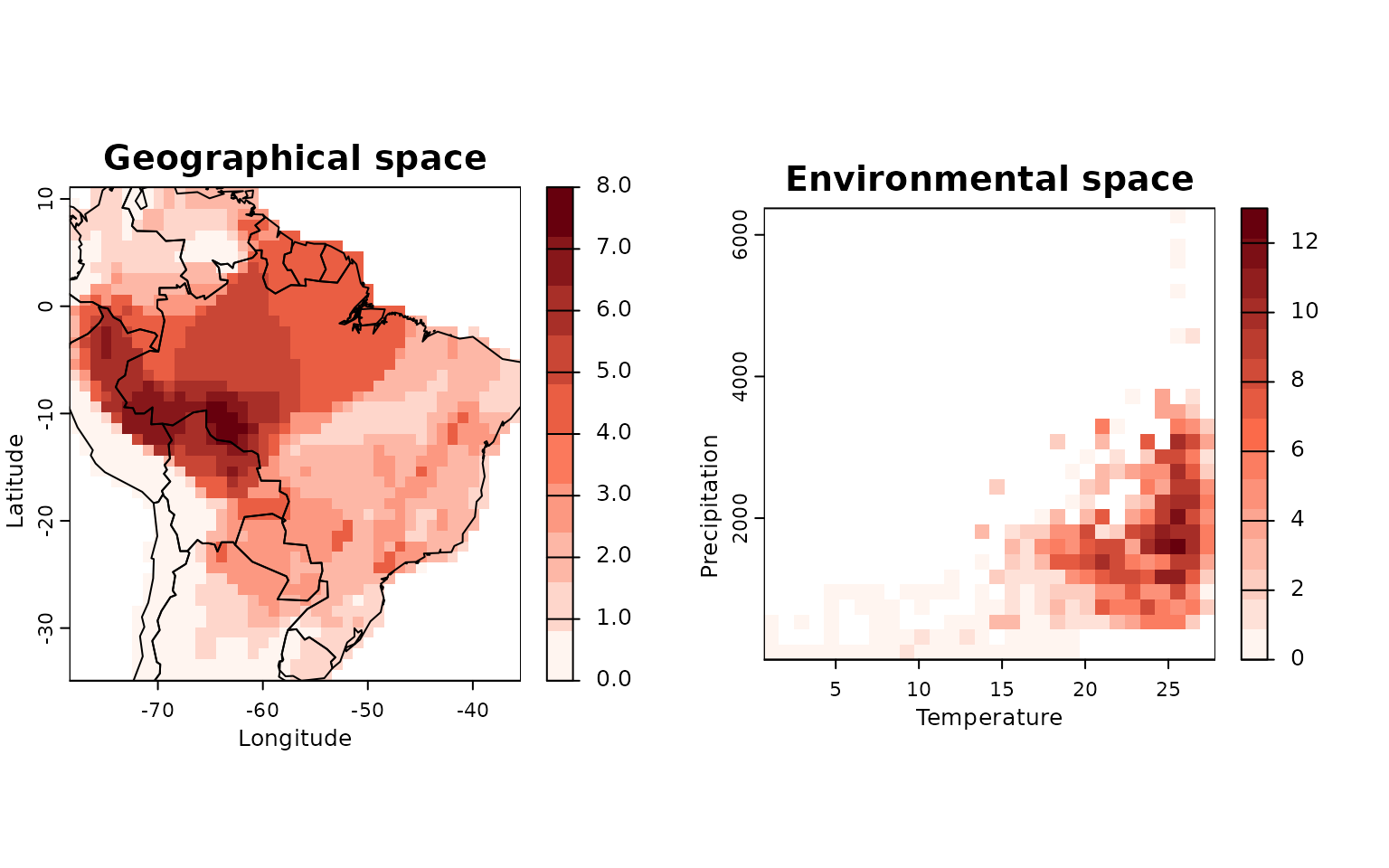
Environmental descriptors per environmental cell
# Compute env-space descriptors (centrality, border, isolation, etc.)
env_cells <- lets.envcells(env_obj, perc = 0.2)
head(env_cells)
#> Cell_env Frequency Isolation (Min.) Isolation (1st Qu.) Isolation (Median)
#> 3 1 0 NA NA NA
#> 4 2 0 NA NA NA
#> 5 3 0 NA NA NA
#> 6 4 0 NA NA NA
#> 7 5 0 NA NA NA
#> 8 6 0 NA NA NA
#> Isolation (Mean) Isolation (3rd Qu.) Isolation (Max.)
#> 3 NA NA NA
#> 4 NA NA NA
#> 5 NA NA NA
#> 6 NA NA NA
#> 7 NA NA NA
#> 8 NA NA NA
#> Weighted Mean Distance to midpoint Mean Distance to midpoint
#> 3 -3.730908 -3.457246
#> 4 -3.646836 -3.382386
#> 5 -3.564523 -3.309862
#> 6 -3.484092 -3.239832
#> 7 -3.405677 -3.172460
#> 8 -3.329421 -3.107918
#> Minimum Zero Distance Minimum 10% Zero Distance Distance to MCP border
#> 3 0 0.9362266 0
#> 4 0 0.8700119 0
#> 5 0 0.8136812 0
#> 6 0 0.7676179 0
#> 7 0 0.7323872 0
#> 8 0 0.7068043 0
#> Frequency Weighted Distance
#> 3 3.800462
#> 4 3.718461
#> 5 3.638213
#> 6 3.559825
#> 7 3.483411
#> 8 3.409092Summarize those per-cell descriptors to species level by aggregating across the environmental cells each species occupies:
# Species-level summaries (e.g., mean across occupied env cells)
env_by_species <- lets.summaryze.cells(env_obj, env_cells, func = mean)
head(env_by_species)
#> Species Frequency Isolation (Min.) Isolation (1st Qu.)
#> 1 Phyllomedusa araguari 9.00000 97052.27 494018.8
#> 2 Phyllomedusa atelopoides 25.38889 109736.78 593524.1
#> 3 Phyllomedusa ayeaye 11.50000 99631.44 384047.4
#> 4 Phyllomedusa azurea 11.59091 130717.35 462970.6
#> 5 Phyllomedusa bahiana 11.91667 136522.70 463613.5
#> 6 Phyllomedusa baltea 19.00000 110574.31 611182.1
#> Isolation (Median) Isolation (Mean) Isolation (3rd Qu.) Isolation (Max.)
#> 1 767952.4 747035.8 923841.2 1500329
#> 2 1109618.7 1175110.9 1714363.8 3008424
#> 3 599271.8 667315.7 851729.1 1725300
#> 4 1391519.0 1328709.9 2016286.4 3035828
#> 5 1492534.7 1354451.4 2121714.6 2999208
#> 6 1019632.7 1201164.4 1896221.5 2950964
#> Weighted Mean Distance to midpoint Mean Distance to midpoint
#> 1 -0.3188860 -0.1627780
#> 2 -0.6007348 -1.0114155
#> 3 -0.3310414 -0.1608610
#> 4 -0.4455612 -0.7009224
#> 5 -0.4520640 -0.6818815
#> 6 -0.6779813 -1.0585809
#> Minimum Zero Distance Minimum 10% Zero Distance Distance to MCP border
#> 1 0.2581989 0.9296124 0.2581989
#> 2 0.3258642 1.0476293 0.3258642
#> 3 0.3116736 0.9521703 0.3116736
#> 4 0.3197515 1.2125076 0.3197515
#> 5 0.2641105 1.2398626 0.2641105
#> 6 0.2581989 0.9893314 0.2581989
#> Frequency Weighted Distance
#> 1 0.7555036
#> 2 0.7967939
#> 3 0.7590072
#> 4 0.7875067
#> 5 0.7752855
#> 6 0.8278369We will use one metric from env_by_species(for example,
“Weighted Mean Distance to midpoint”) for cross-mapping.
Attribute-space PAM for Phyllomedusa species
We create a trait grid (attribute space) by simulating two quantitative traits for the species present in our PAM. (Replace with real traits if available.)
set.seed(123)
sp_vec <- env_by_species$Species # species present in PAM
n_sp <- length(sp_vec)
trait_a <- rnorm(n_sp)
trait_b <- trait_a * 0.2 + rnorm(n_sp) # correlated trait
attr_df <- data.frame(Species = sp_vec,
trait_a = trait_a,
trait_b = trait_b)
# Attribute-space PAM (AttrPAM)
attr_obj <- lets.attrpam(attr_df, n_bins = 5, remove.cells = FALSE)
# Richness map in attribute space
lets.plot.attrpam(attr_obj)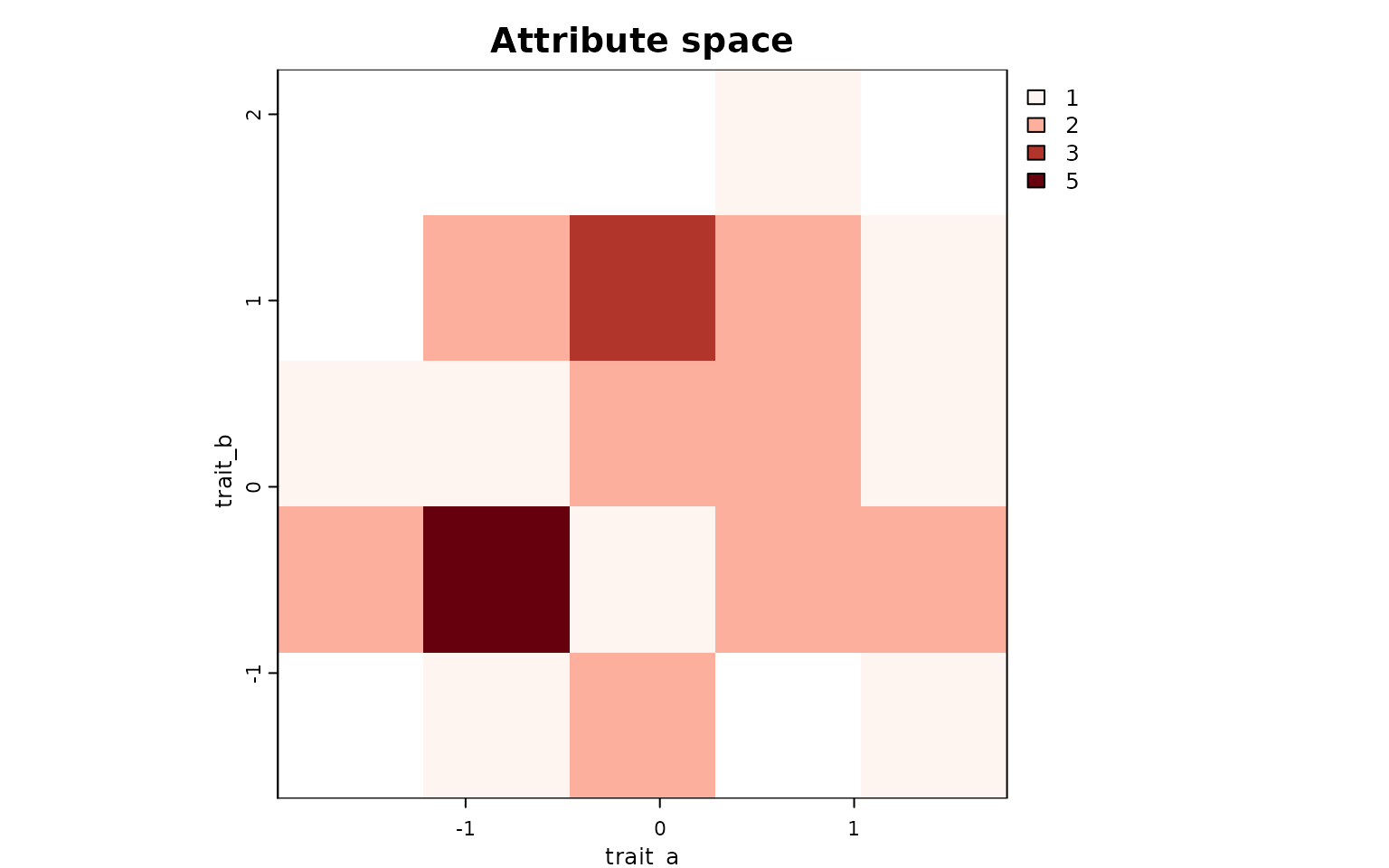
Cross-mapping an environmental metric into geographic and attribute spaces
A. Into geographic space
Project environmental metric to geography:
# Align env-cell descriptors to the order of geographic rows
env_cells_geo <- env_cells[ env_obj$Presence_and_Absence_Matrix_geo[, 1], ]
# Template = geographic richness raster
map_richatt2 <- env_obj$Geo_Richness_Raster
terra::values(map_richatt2) <- NA
# Fill geographic cells with the env-space metric (species-level NOT needed here)
terra::values(map_richatt2)[ env_obj$Presence_and_Absence_Matrix_geo[, 2] ] <-
env_cells_geo$`Weighted Mean Distance to midpoint`
# Palette and plot
colfunc <- grDevices::colorRampPalette(c("#fff5f0", "#fb6a4a", "#67000d"))
plot(map_richatt2, col = colfunc(200), box = FALSE, axes = FALSE,
main = "Env centrality (env-space) mapped to geography")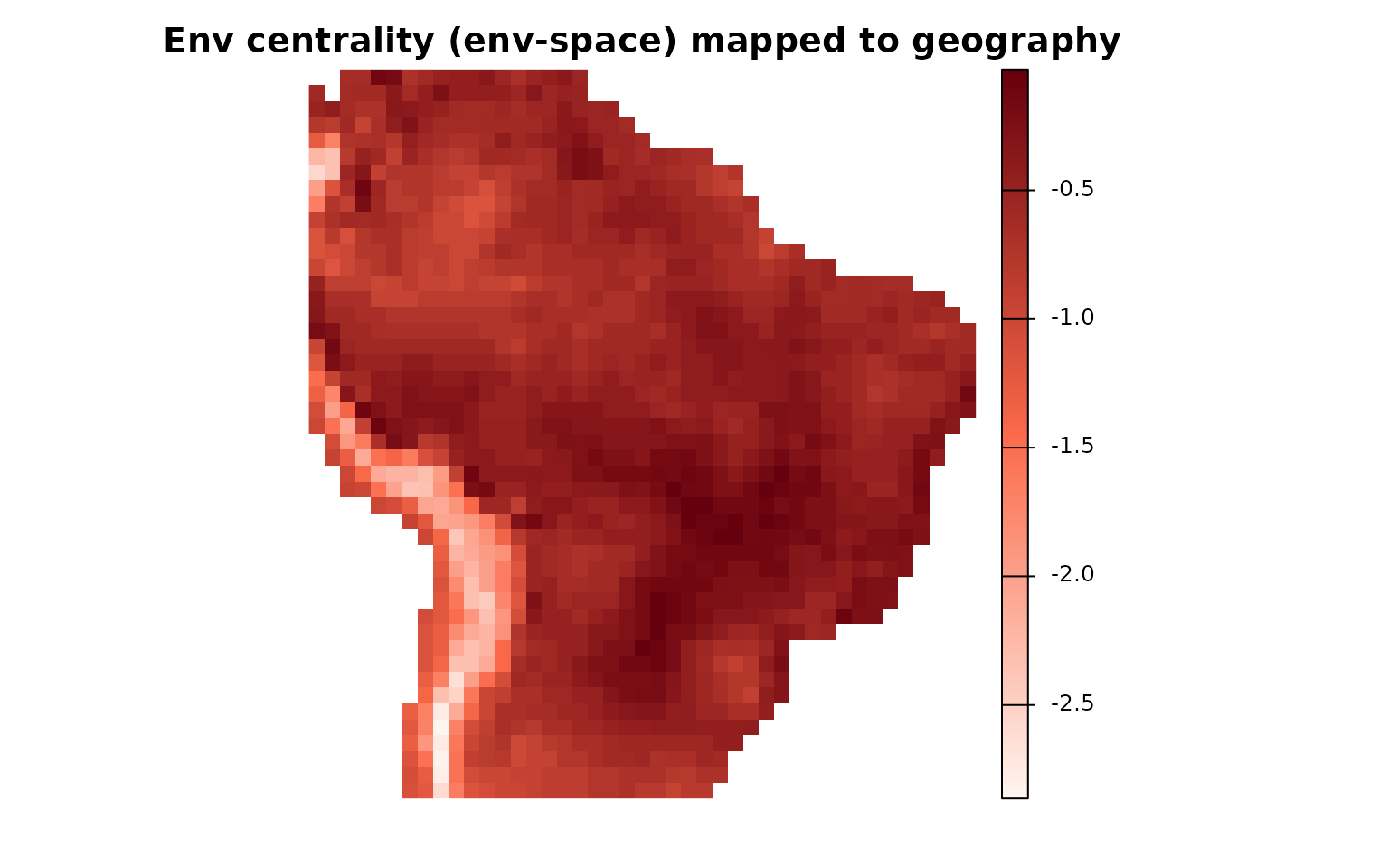
B. Into attribute space
Map the same environmental metric (species-level) into the attribute grid:
met_env <- env_by_species$`Weighted Mean Distance to midpoint`
sp_names <- env_by_species$Species
attr_map <- lets.maplizer.attr(attr_obj, y = met_env, z = sp_names, func = mean)
# Visualize
lets.plot.attrpam(attr_map, mar = c(4,4,4,4))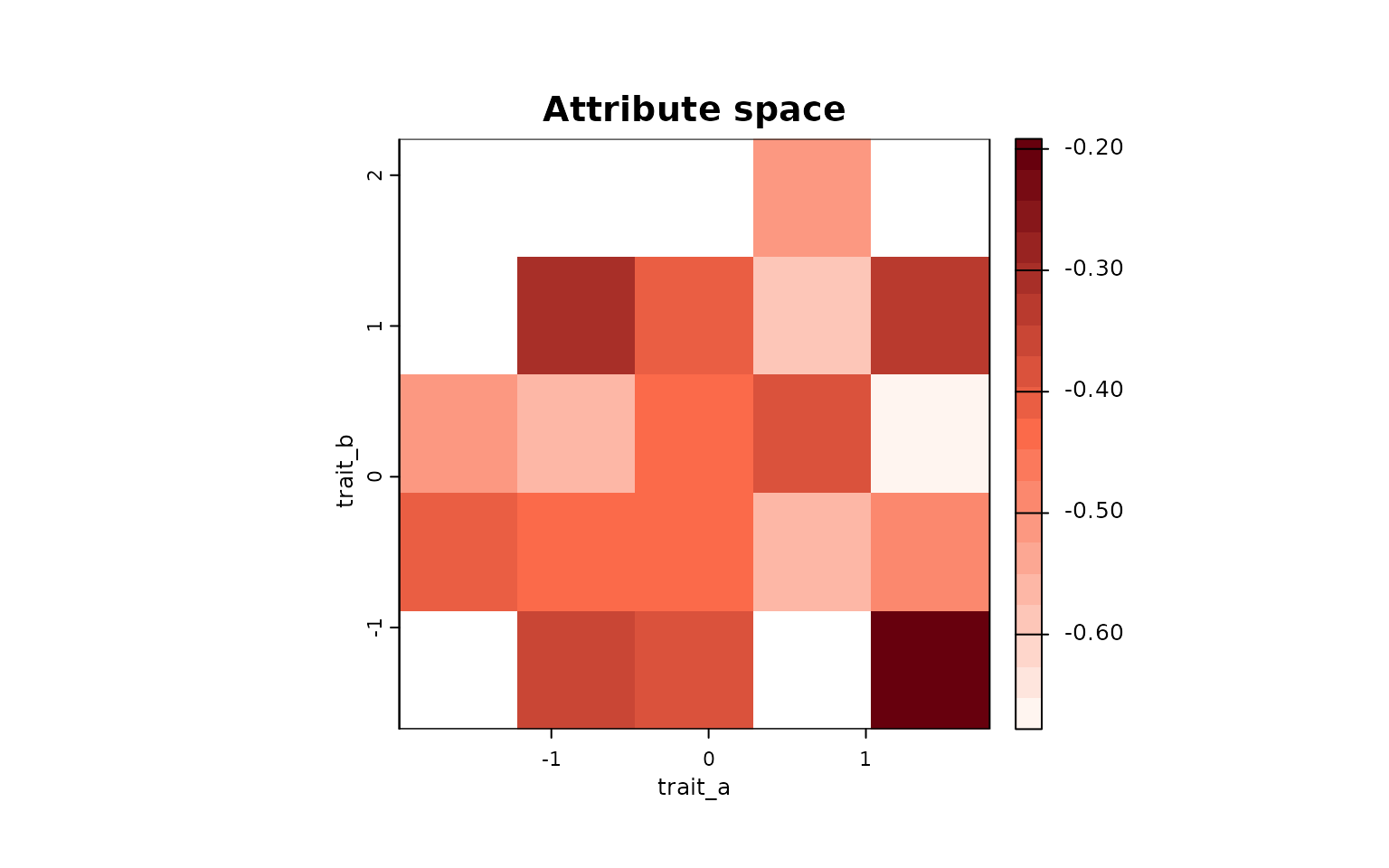
These projections reveal how a descriptor computed in environmental space (centrality) distributes across geographic communities and trait space.
Cross-mapping attribute metrics back to geography
You can compute attribute-space descriptors with
lets.attrcells(attr_obj, ...), summarize them to species
with lets.summarizer.cells(attr_obj, ...), and then project
those species-level metrics to geographic or environmental spaces using
lets.maplizer(...) or
lets.maplizer.env(...).
# Attribute-space descriptors per cell
attr_cells <- lets.attrcells(attr_obj, perc = 0.2)
# Species-level summaries of attribute-space descriptors
attr_by_species <- lets.summaryze.cells(attr_obj, attr_cells, func = mean)
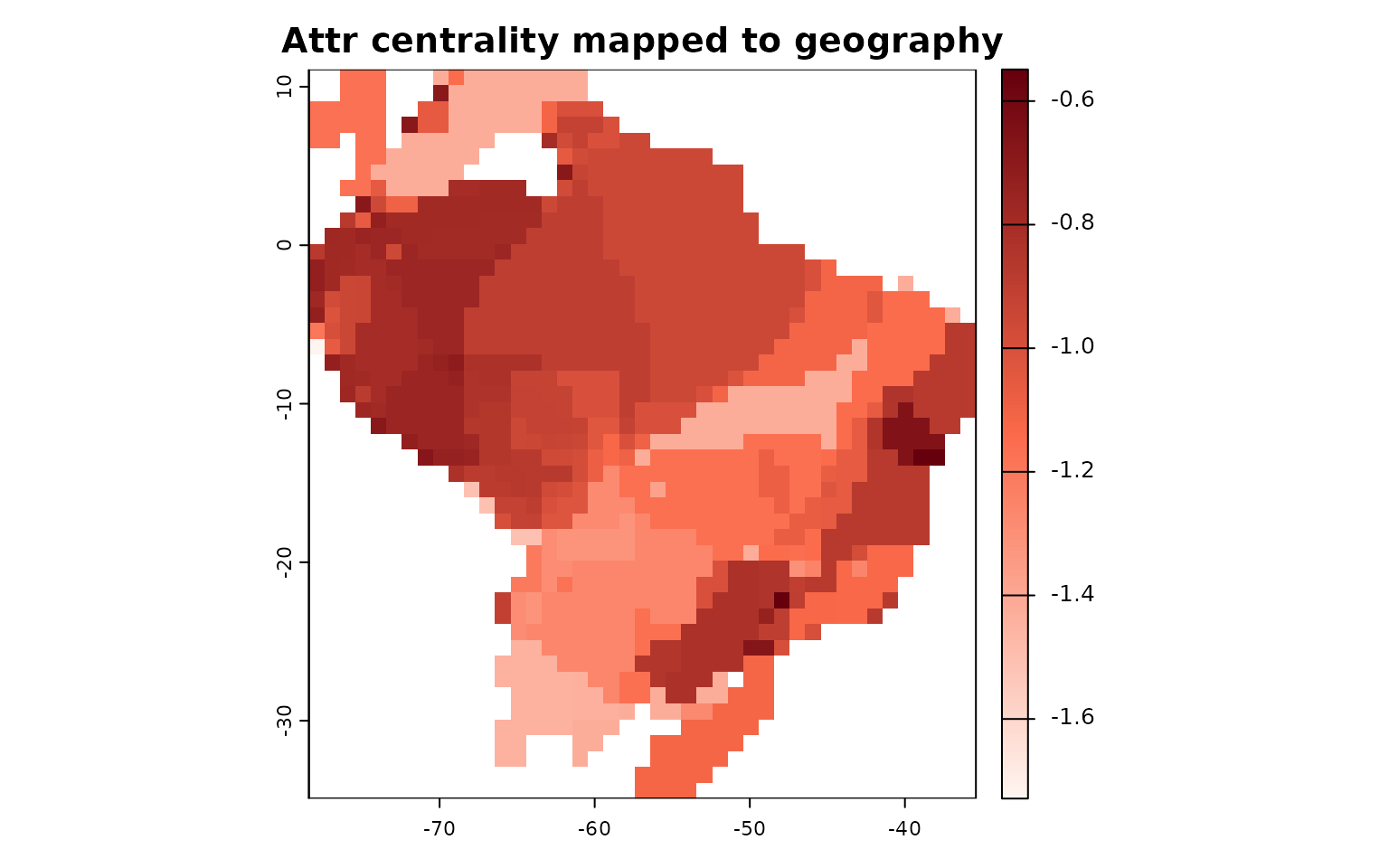
# Example: map an attribute-space centrality back to geography
met_attr <- attr_by_species$`Weighted Mean Distance to midpoint`
geo_from_attr <- lets.maplizer(PAM, y = met_attr, z = attr_by_species$Species, ras = TRUE)
plot(geo_from_attr$Raster, main = "Attr centrality mapped to geography", col = colfunc(200))